Adrenergic Receptors
Functional (agonist/antagonist) readouts for α₁D were not reported in the same studies; however, the binding Kᵢ pattern
and the α₁D pharmacology suggest antagonist-like behavior would be consistent with the structural class (yohimbine-like)
and with α₁ antagonists’ canonical effects. This is a cautious inference; direct functional α₁D data for corynantheidine
are not yet published in the sources cited here.
In Vivo Nociception
Mouse antinociception (i.c.v., warm-water tail withdrawal): corynantheidine produced a ceiling ~50% MPE, and the effect
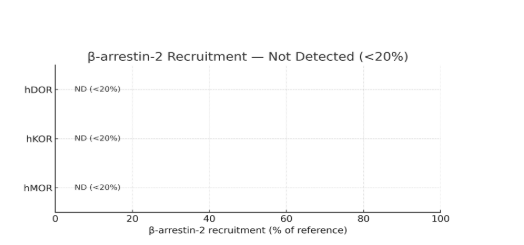
was MOR-dependent in MOR-KO mice, consistent with MOR partial agonism and absence of β-arrestin recruitment [12].
Structure–Activity Context (SAR)
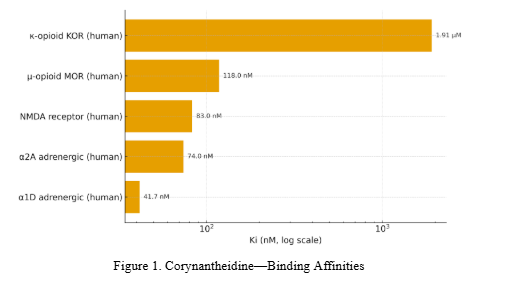
C-9 methoxy removal (mitragynine → corynantheidine) does not reduce MOR binding but reduces KOR affinity; docking
attributes this to loss of a methoxy–Thr111 interaction in KOR and steric/hydrogen-bonding differences across subpockets [13].
Earlier docking/structure work on mitragynine-template ligands supports key residue roles (e.g., Trp293, His297, Gln124, Tyr128)
that rationalize MOR/KOR differences [14].
Serotonergic Evidence
For corynantheidine, no validated 5-HT receptor binding/function has been reported in the sources above.
(Recent serotonergic assays on kratom alkaloids emphasized other congeners—e.g., speciogynine, mitragynine—rather
than corynantheidine.) Any serotonergic role for corynantheidine remains unconfirmed in peer-reviewed primary data [15].
Evidence Limits and Requirements
- Serotonergic: No validated 5-HT binding/functional data for corynantheidine in the primary sources; serotonergic activity is established for other kratom indoles (e.g., paynantheine at 5-HT₁A/5-HT₂B), not for corynantheidine. Do not generalize across congeners [16].
- Adrenergic function: α₁D binding is robust; functional α₁D (agonism/antagonism) for corynantheidine is not yet published in the cited datasets; inference of α₁D antagonism remains a hypothesis until tested [17].
- Species/assay: hMOR (BRET) vs mMOR ([³⁵S]GTPγS) differ in apparent efficacy; report species, cell line, and assay for all comparisons [18].
Practical Implications
The profile differs from potent MOR agonists: partial MOR agonism, no β-arrestin-2, and high α₁D binding.
If exposures at target tissues are sufficient, α₁D engagement could contribute non-opioid effects
(e.g., vascular or smooth muscle modulation); this needs direct functional testing (Ca²⁺ flux or IP₁ assays)
at α₁D and human PK to relate in vitro potency to feasible in vivo concentrations [19].